Interfaces to search engines
I have shown previously some unique aspects in exporting PSMs from Mascot, for uses with proteoQ. In this post, I will do the same for more search engines, e.g. MaxQuant and MSFragger.
MaxQuant
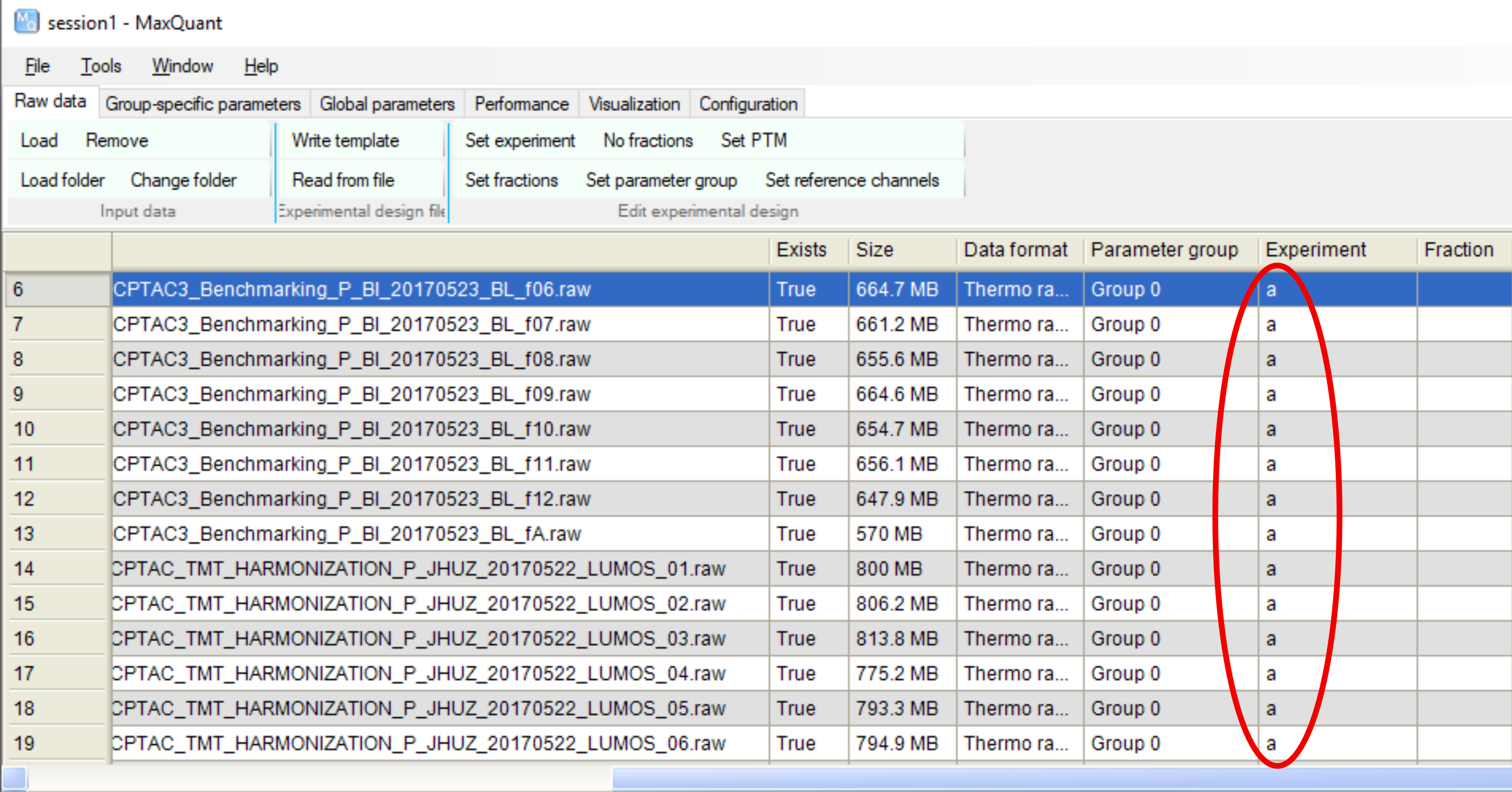
When performing MaxQuant searches, we would be asked to provide the names of experiments, and cannot simply leave them blank. One immediate question is: how would these names be related to the information in the metadata files (i.e. expt_smry.xlsx and frac_smry.xlsx) in proteoQ?
As you may have sorted out, the additional knowledge about the names of experiments will not be consumed with proteoQ. In the example of TMT experiments, connections among peptide spectrum matches (PSM), sample IDs and TMT channels are fully established with the metadata in the two .xlsx files mentioned above. Such certainty would hold for LFQ workflows as well. To do as little as needed, we may simply enter some dummy names when running MaxQuant:
MSFragger
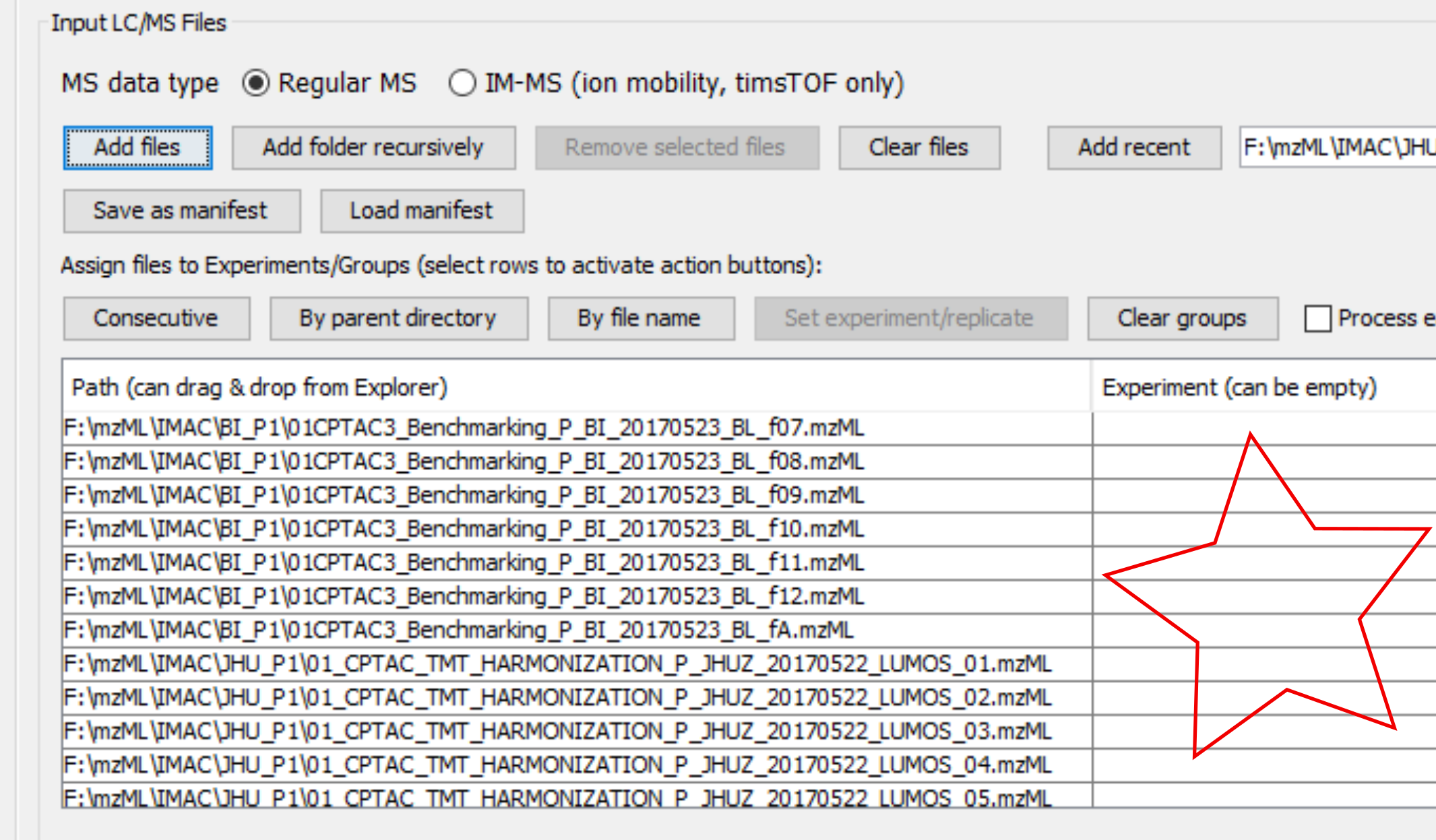
Analogously in the settings of MSFragger searches, we may leave the fields under Experiment empty, for both TMT and LFQ searches.
In case of TMT procedures, we will be further prompted to compile a lookup between TMT channels and sample IDs. We may again enter dummy names, regardless that there are one or multiple TMT experiments being fed to the program:
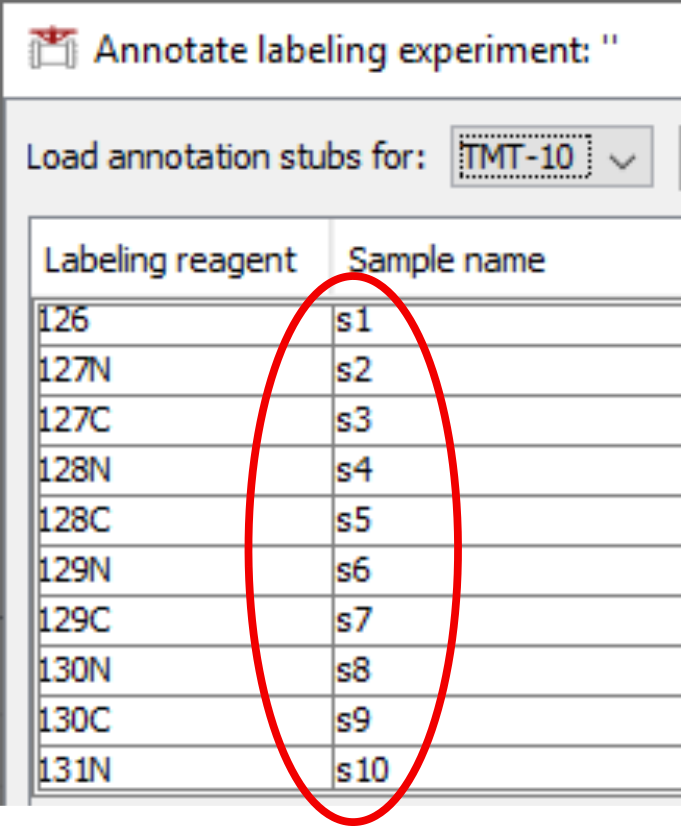
If we want to be too savvy in spending our time, we may simply named them identically, e.g. s, and let the program add some differentiating suffices for us.
Multiple experiments at once
For both MaxQuant and MSFragger, the approaches described in the post are fully compatible for a merged search with multiple TMT experiments. We don’t need to run one search at a time. In other words, the procedures are irrespective to the number of TMT experiments. We would not incur ambiguities such as the same dummy name across different TMT plexes. The unique correspondences between sample IDs and experiment indexes, as well as TMT channels, can be unambiguously established with the information in expt_smry.xlsx and frac_smry.xlsx.
The same is true for LFQ workflows.